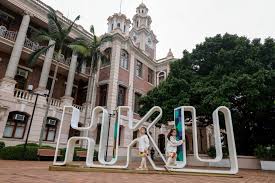Postdoctoral and Research Positions at KTH Royal Institute of Technology in Sweden, a workplace that has a...
Postdoctoral Scholarships at Karolinska Institutet in Sweden, a research-led medical university in Solna within the Stockholm urban...
PhD Scholarships at Karolinska Institutet, a research-led medical university in Solna within the Stockholm urban area of...
Postdoctoral Scholarships at Chalmers University of Technology, a Swedish university located in Gothenburg, Sweden. Postdoctoral Scholarships at...
PhD Scholarships at Chalmers University of Technology, a Swedish university located in Gothenburg, Sweden. PhD Scholarships at...
Lund University, consistently ranked among the top institutions in Northern Europe, is actively recruiting for approximately 20...
PhD Scholarships at Lund University in Sweden, one of northern Europe’s oldest universities in Sweden There are...
The University of Hong Kong invites application for vacant Teaching and Faculty Positions, a public research university...
The University of Hong Kong invites application for vacant Teaching and Faculty Positions, a public research university...
City University of Hong Kong invites application for vacant Postdoc and Research Positions, a public research university...